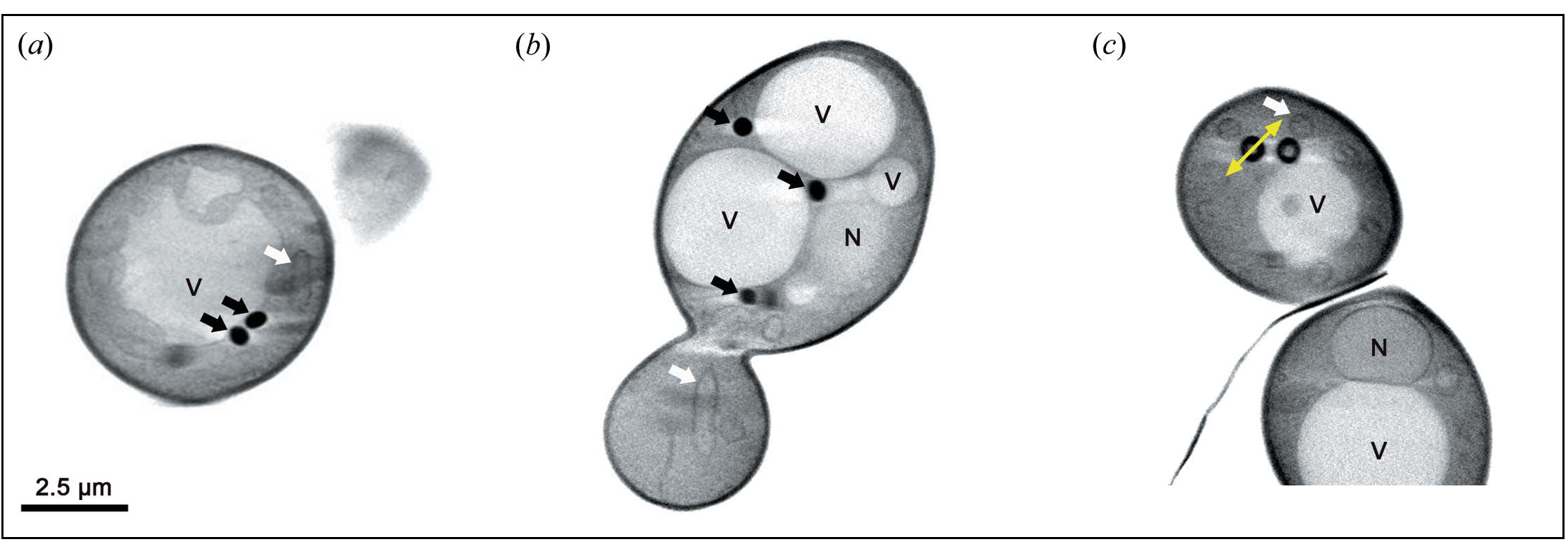
Lipid droplets core heterogeneity revealed by cryo soft-X-rays tomography. Images obtained with the transmission X-ray microscope of MISTRAL at ALBA. Nucleus (N), vacuole (V), lipid droplet (black arrow) and mitochondria (white arrow).
Cerdanyola del Vallès, 21st May 2020 A new non-invasive cell imaging method has been developed by adapting and combining two imaging techniques previously used to study plant and animal cells. In this case, this new method was employed to study cells of baker's yeast (Saccharomyces cerevisiae) and its lipid droplets.
Lipid droplets are one of the organelles that compose the cells of the complex organisms. They have been the focus of intense study because of their key role in several diseases, including diabetes and obesity. The lipids found in the droplets are of interest to the food industry (e.g., for making vegetable oils), the green chemistry industry (e.g., for making biodiesel), and the cosmetics industry (e.g., for making soaps and lotions containing vegetable oils). Diverse lipids are present within the droplets, although triglycerides and cholesterol esters predominate and are responsible for droplet structure. Thus, clarifying the structure and composition of lipid droplets has important practical consequences.
Current methods for studying the internal structure of lipid droplets destroy cells. These methods have led to major discoveries about the structure and composition of individual droplets or droplets observed in sectioned cells. It is now necessary to develop new imaging tools to more comprehensively study how the droplets function and interact with other organelles within intact cells.
In this study, researchers used cryo soft X-ray tomography, at the MISTRAL beamline of the ALBA Synchrotron, to analyse yeast cells exposed to ultrarapid freezing (i.e., vitrified yeast cells). This X-ray-based technique makes it possible to study the internal architecture of cells at the nanometric scale (one billionth of a metre). More specifically, the technique can reveal organelle arrangements and interactions. Lipid droplets strongly absorb X-rays and can thus be clearly seen in the resulting images.
Secondly, researchers used deep-UV imaging (employed at the SOLEIL synchrotron). This technique does not require any special preliminary procedures and allows researchers to observe living cells at micrometric to nanometric scales. Consequently, it can be used to observe the dynamics of internal cellular processes.
Revealing the internal structure of cells ten times smaller than plant or animal cells
The challenge was to adapt this technique for use on yeast cells, which are 10 times smaller than plant or animal ones. Cryo soft X-ray tomography performed at MISTRAL showed that the structure of lipid droplets changed depending on their composition. Deep-UV imaging in SOLEIL provided information about the structure of and contact between organelles in a living yeast cell at a scale of around 100 nanometres, without the need for any chemical markers. The researchers also developed specific procedures for combining different UV imaging techniques (i.e., based on the transmittance and fluorescence of tryptophan and tyrosine, two amino acids) with a view to studying living cells.
By using cryo soft X-ray tomography and deep-UV imaging in tandem, the researchers were able to obtain complementary information on cells at the micrometric to nanometric scale. The x-ray-based imaging technique can be used to explore the detailed structure of cell organelles. In contrast, the UV-based imaging technique can be used to characterise biological processes within cells (e.g., cell division); determine the fate of molecules that originate from outside the cell; and examine cellular responses to stress. This new non-invasive cell imaging method could also be used to analyse other types of cells, notably plant or animal cells, and could thus prove beneficial to other synchrotron researchers and biologists in general. The method will make it easier to build a new organelle atlas in living cells.
Reference: Frédéric Jamme, Bertrand Cinquin, Yann Gohon, Eva Pereiro, Matthieu Réfrégiers et Marine Froissard, Synchrotron multimodal imaging in a whole cell reveals lipid droplet core organization, Journal of Synchrotron radiation, May 2020. DOI: https://doi.org/10.1107/S1600577520003847




